1. Getting started
1.1. Basic functions
fsave <- function(x, file, location = "./data/processed/", ...) {
if (!dir.exists(location))
dir.create(location)
datename <- substr(gsub("[:-]", "", Sys.time()), 1, 8)
totalname <- paste(location, datename, file, sep = "")
print(paste("SAVED: ", totalname, sep = ""))
save(x, file = totalname)
}
fpackage.check <- function(packages) {
lapply(packages, FUN = function(x) {
if (!require(x, character.only = TRUE)) {
install.packages(x, dependencies = TRUE)
library(x, character.only = TRUE)
}
})
}
colorize <- function(x, color) {
sprintf("<span style='color: %s;'>%s</span>", color, x)
}1.2. Packages
packages = c("RsienaTwoStep", "doParallel", "ggplot2")
fpackage.check(packages)
#> Loading required package: RsienaTwoStep
#> Loading required package: foreach
#> Loading required package: doParallel
#> Warning in library(package, lib.loc = lib.loc, character.only = TRUE,
#> logical.return = TRUE, : there is no package called 'doParallel'
#> Installing package into '/home/runner/work/_temp/Library'
#> (as 'lib' is unspecified)
#> also installing the dependencies 'listenv', 'parallelly', 'future', 'globals', 'shape', 'future.apply', 'numDeriv', 'progressr', 'SQUAREM', 'diagram', 'lava', 'tzdb', 'cpp11', 'prodlim', 'timechange', 'proxy', 'Rcpp', 'data.table', 'clock', 'gower', 'hardhat', 'ipred', 'lubridate', 'timeDate', 'e1071', 'ModelMetrics', 'plyr', 'pROC', 'recipes', 'reshape2', 'caret', 'mlbench', 'RUnit'
#> Loading required package: iterators
#> Loading required package: parallel
#> Loading required package: ggplot2
#> [[1]]
#> NULL
#>
#> [[2]]
#> [1] "doParallel" "parallel" "iterators" "RsienaTwoStep"
#> [5] "foreach" "stats" "graphics" "grDevices"
#> [9] "utils" "datasets" "methods" "base"
#>
#> [[3]]
#> NULL2. Setting up cluster
no_cores <- detectCores() - 1
mycl <- makeCluster(rep("localhost", no_cores))
clusterEvalQ(mycl, library(RsienaTwoStep))
registerDoParallel(mycl)
#stopCluster(mycl)3. Running simulations on toy data
3.1. Model 1: Degree and reciprocity
Let us assume people really don’t like to have a non-reciprocal tie but do like reciprocal ties a lot. (I only want to help you if you help me!)
Thus if you start with a network without many (reciprocal) ties it would be very difficult to get more reciprocal ties in the normal model. However, with simultaneity this should be possible.
3.1.1. Simulate networks
NSIM <- 500
STATS <- list(ts_degree, ts_recip)
STARTS <- c(10, -1,2)
sims1 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(1,0,0), verbose = FALSE, parallel = TRUE) #ministep
sims2 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), verbose = FALSE, parallel = TRUE) #twostep: simultaneity
sims3 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), dist1 = 2, dist2 = 2, verbose = FALSE, parallel = TRUE) #twostep: weak coordination
sims4 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), dist1 = 2, verbose = FALSE, parallel = TRUE) #twostep: strict coordination
sims5 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,0,1), dist1 = 2, verbose = FALSE, parallel = TRUE) #simstep: one actor two ministeps3.1.2. Counting dyads
df1 <- ts_dyads(sims=sims1, simtype="ministep")
df2 <- ts_dyads(sims=sims2, simtype="twostep: simultaneity")
df3 <- ts_dyads(sims=sims3, simtype="twostep: weak coordination")
df4 <- ts_dyads(sims=sims4, simtype="twostep: strict coordination")
df5 <- ts_dyads(sims=sims5, simtype="simstep")
df <- rbind(df1, df2, df3, df4, df5)3.1.3. Plot results of dyadcensus
p <- ggplot(df, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep")))) +
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "dyad type", y = "dyad count", fill="simulation type")
p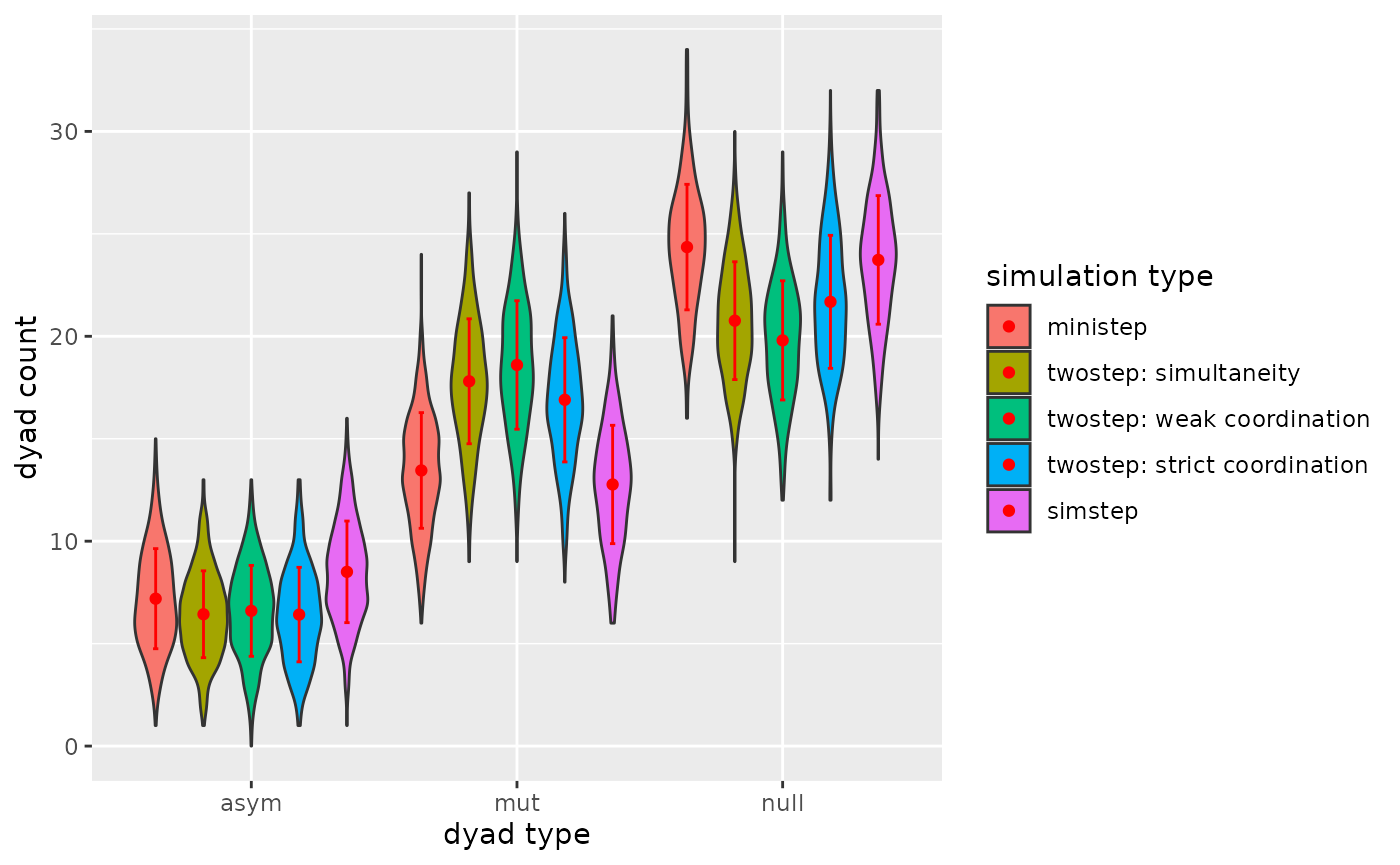
Figure 3.1.3. Dyadcensus Model 1
3.1.4. Conclusion
Yes, if you allow simultaneity, we will observe more reciprocal ties after same number of possible tie-changes.
Perhaps the model with ministeps only would reach the same target values (for dyads) but probably by using more possible tie-changes. This would mean there will be more noise in the network chains, this can/will have results for other network statistics.
Also note, that if you make the network larger, simultaneity will less likely to be different from two consecutive ministeps, this is because the two agents do not influence each others evaluation functions.
3.2. Model 2: Degree, reciprocity, transTrip
Let us assume people really don’t like to have a non-reciprocal tie but do like reciprocal ties a lot and do favor transitivity.
Is it unlikely that a transitive triad will emerge if we have coordination, because for one of the actors will not evaluate the transitive triad positively because in wrong position. Or, because reciprocal ties are more likely when actors coordinate, we will also observe more transitive triads.
3.2.1. Simulate networks
NSIM <- 500
STATS <- list(ts_degree, ts_recip,ts_transTrip)
STARTS <- c(10, -1, 2, 1)
sims1 <- ts_sims(startvalues = STARTS , net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(1,0,0), verbose = FALSE, parallel = TRUE) #ministep
sims2 <- ts_sims(startvalues = STARTS , net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), verbose = FALSE, parallel = TRUE) #twostep: simultaneity
sims3 <- ts_sims(startvalues = STARTS , net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), dist1 = 2, dist2 = 2, verbose = FALSE, parallel = TRUE) #twostep: weak coordination
sims4 <- ts_sims(startvalues = STARTS , net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), dist1 = 2, verbose = FALSE, parallel = TRUE) #twostep: strict coordination
sims5 <- ts_sims(startvalues = STARTS , net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,0,1), dist1 = 2, verbose = FALSE, parallel = TRUE) #simstep: one actor two ministeps3.2.2. Counting dyads
df1 <- ts_dyads(sims=sims1, simtype="ministep")
df2 <- ts_dyads(sims=sims2, simtype="twostep: simultaneity")
df3 <- ts_dyads(sims=sims3, simtype="twostep: weak coordination")
df4 <- ts_dyads(sims=sims4, simtype="twostep: strict coordination")
df5 <- ts_dyads(sims=sims5, simtype="simstep")
df <- rbind(df1, df2, df3, df4, df5)3.2.3. Plot results of the dyadcensus
p <- ggplot(df, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep")))) +
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "dyad type", y = "dyad count", fill="simulation type")
p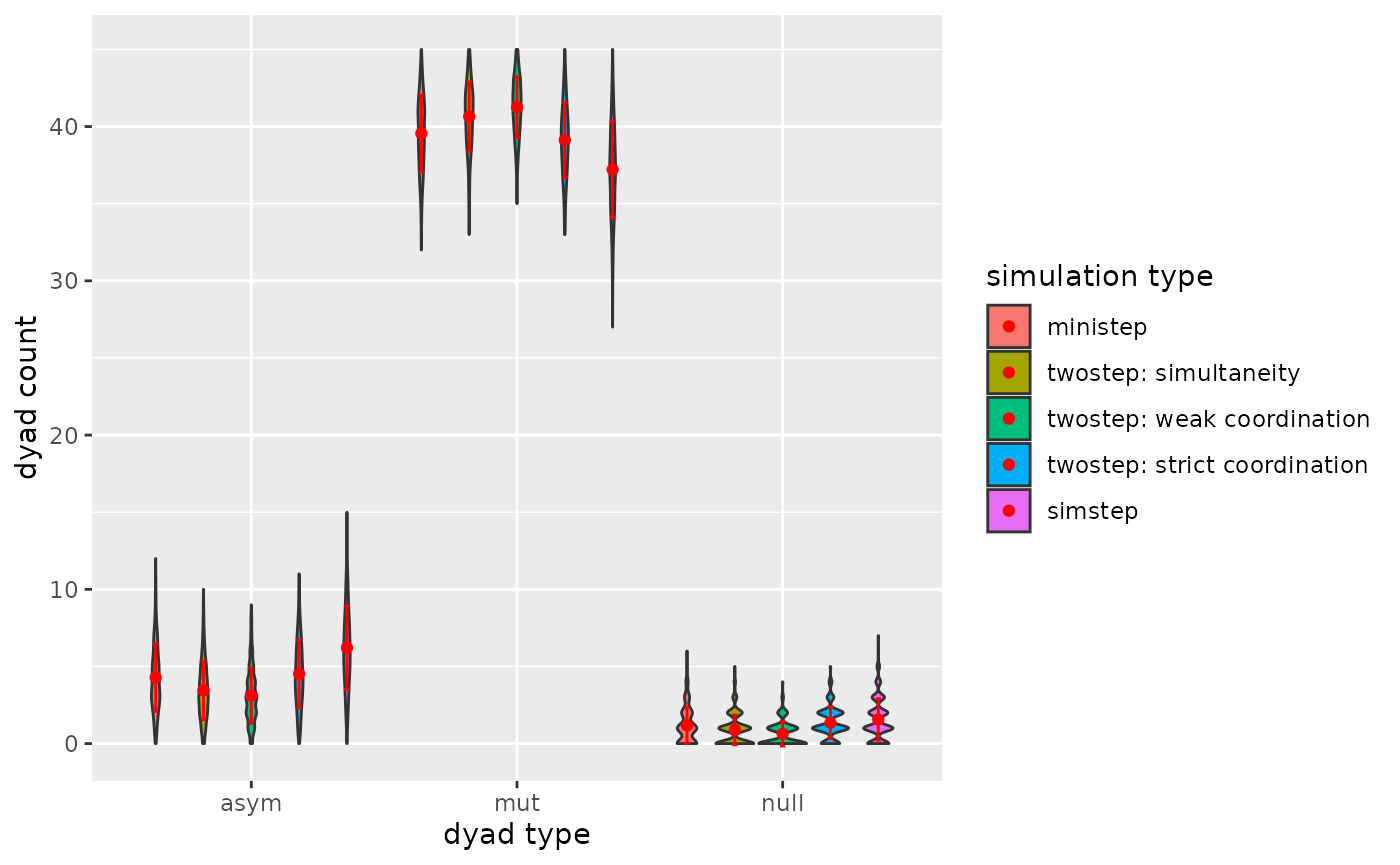
Figure 3.2.3. Dyadcensus Model 2
3.2.4. Counting triads
df1 <- ts_triads(sims=sims1, simtype="ministep")
df2 <- ts_triads(sims=sims2, simtype="twostep: simultaneity")
df3 <- ts_triads(sims=sims3, simtype="twostep: weak coordination")
df4 <- ts_triads(sims=sims4, simtype="twostep: strict coordination")
df5 <- ts_triads(sims=sims5, simtype="simstep")
df <- rbind(df1, df2, df3, df4, df5)3.2.5. Plot results of the triadcensus
Intermezzo
The different triad configurations may embed one or more transitive triads. This is my quick count:
- 030T: 1
- 120U: 1
- 120D: 2
- 120C: 2
- 210: 4
- 300: 6
dftest <- df[df$x=="030T" | df$x=="120U", ]
p1 <- ggplot(dftest, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep")))) +
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "triad type", y = "triad count", fill="simulation type")
p1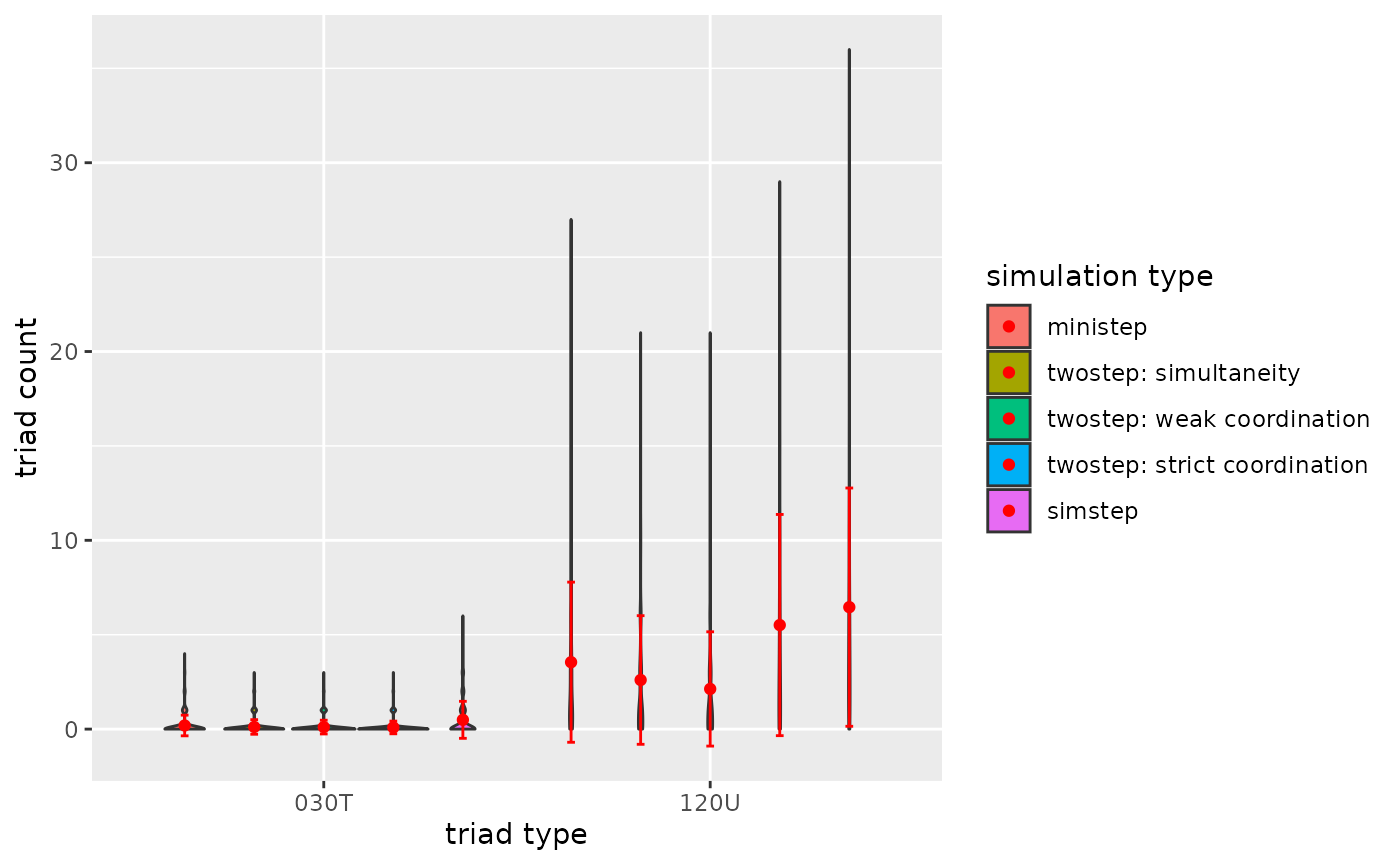
Figure 3.2.5a. Triadcensus Model 2 (Triads 030T & 120U)
dftest <- df[df$x=="120D" | df$x=="120C" , ]
p2 <- ggplot(dftest, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep")))) +
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "triad type", y = "triad count", fill="simulation type")
p2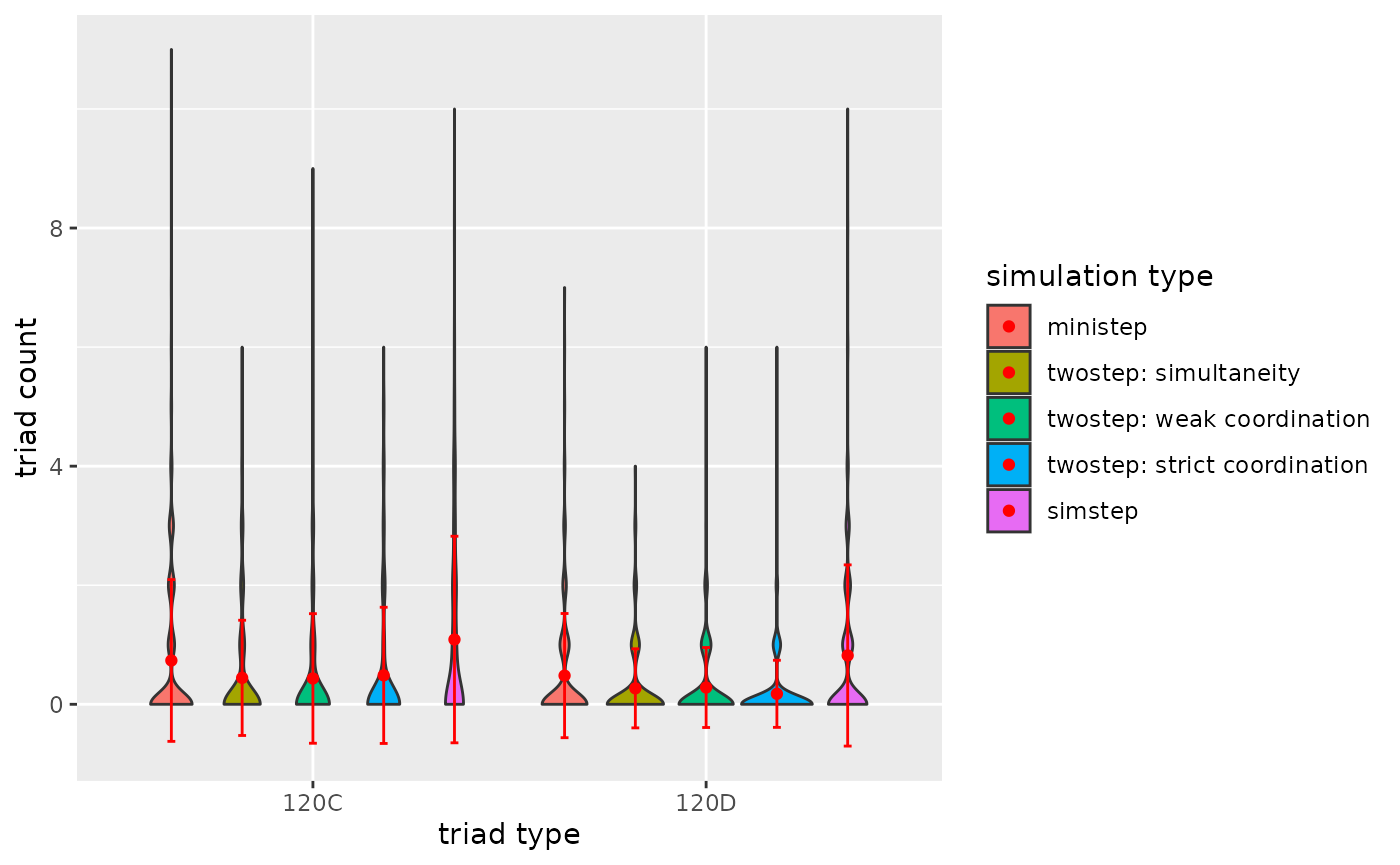
Figure 3.2.5b. Triadcensus Model 2 (Triads 120D & 120C)
dftest <- df[df$x=="210" | df$x=="300" , ]
p3 <- ggplot(dftest, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep")))) +
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "triad type", y = "triad count", fill="simulation type")
p3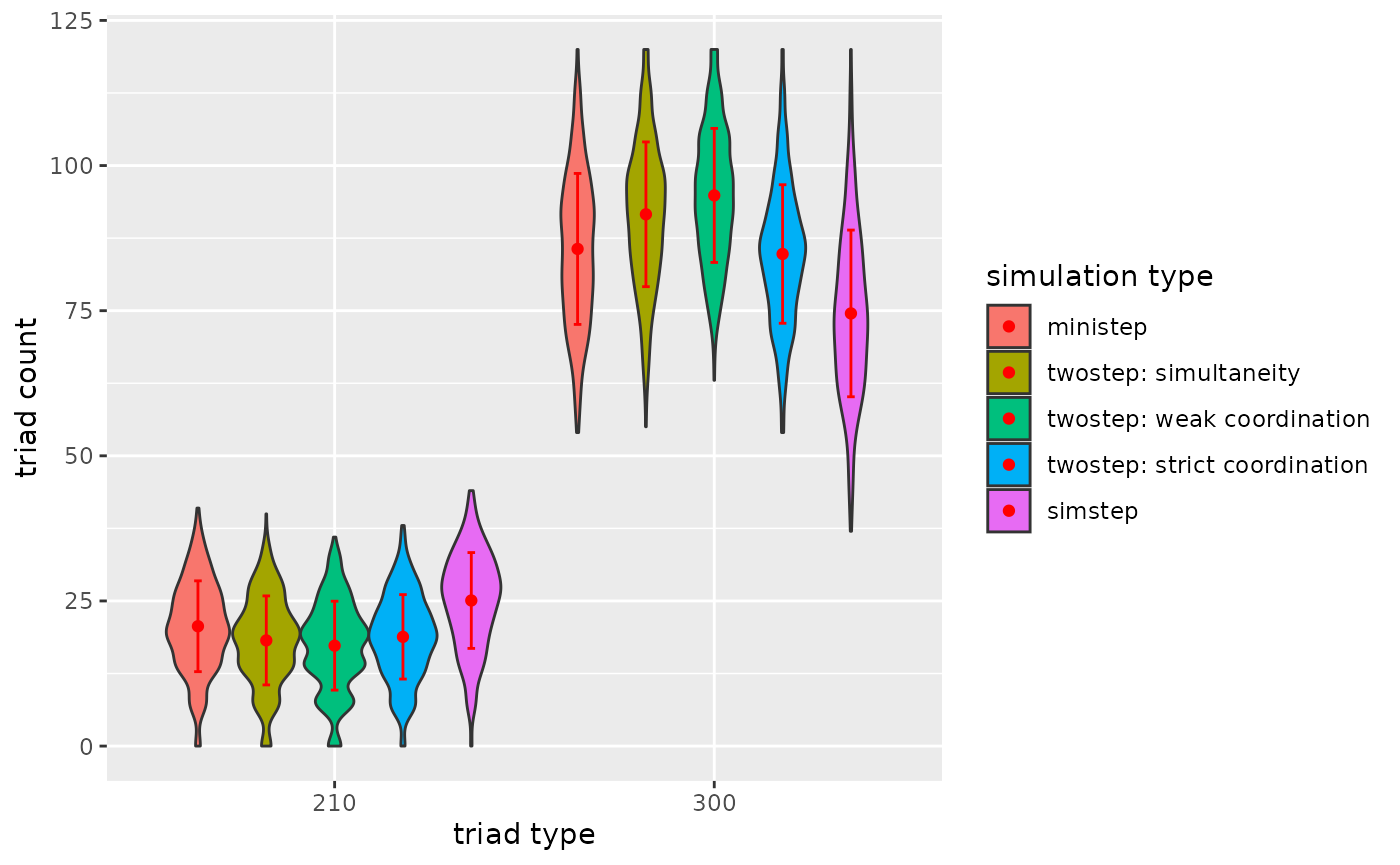
Figure 3.2.5c. Triadcensus Model 2 (Triads 210 & 300)
3.2.6. Conclusion
For this network (ts_net1) and given the network
statistics degree, reciprocity and transitivity…
- A general conclusion is that the type of ABM does impact the dyad
and triad count after the simulation run.
- A tentative conclusion would be that weak coordination impacts (complex) triad configurations more than strict coordination, because under weak coordination asymmetric dyads are plausible.
3.3. Model 3: Degree, reciprocity, transTrip, transMedTrip
Let us assume people really don’t like to have a non-reciprocal tie but do like reciprocal ties a lot and do favor transitivity.
It is now likely that a transitive triad will emerge if we have coordination, because one of the actors evaluate the transTrip and the other may positively evaluate transMedTrip positively.
3.3.1. Simulate networks
NSIM <- 500
STATS <- list(ts_degree, ts_recip, ts_transTrip, ts_transMedTrip)
STARTS <- c(10, -2 ,1 ,2 ,2)
sims1 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(1,0,0), verbose = FALSE, parallel = TRUE) #ministep
sims2 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), verbose = FALSE, parallel = TRUE) #twostep: simultaneity
sims3 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), dist1 = 2, dist2 = 2, verbose = FALSE, parallel = TRUE) #twostep: weak coordination
sims4 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,1,0), dist1 = 2, verbose = FALSE, parallel = TRUE) #twostep: strict coordination
sims5 <- ts_sims(startvalues = STARTS, net = ts_net1, statistics = STATS, nsims = NSIM, p2step = c(0,0,1), dist1 = 2, verbose = FALSE, parallel = TRUE) #simstep: one actor two ministeps3.3.2. Counting dyads
df1 <- ts_dyads(sims=sims1, simtype="ministep")
df2 <- ts_dyads(sims=sims2, simtype="twostep: simultaneity")
df3 <- ts_dyads(sims=sims3, simtype="twostep: weak coordination")
df4 <- ts_dyads(sims=sims4, simtype="twostep: strict coordination")
df5 <- ts_dyads(sims=sims5, simtype="simstep")
df <- rbind(df1, df2, df3, df4, df5)3.3.3. Plot results of the dyadcensus
p <- ggplot(df, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep")))) +
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "dyad type", y = "dyad count", fill="simulation type")
p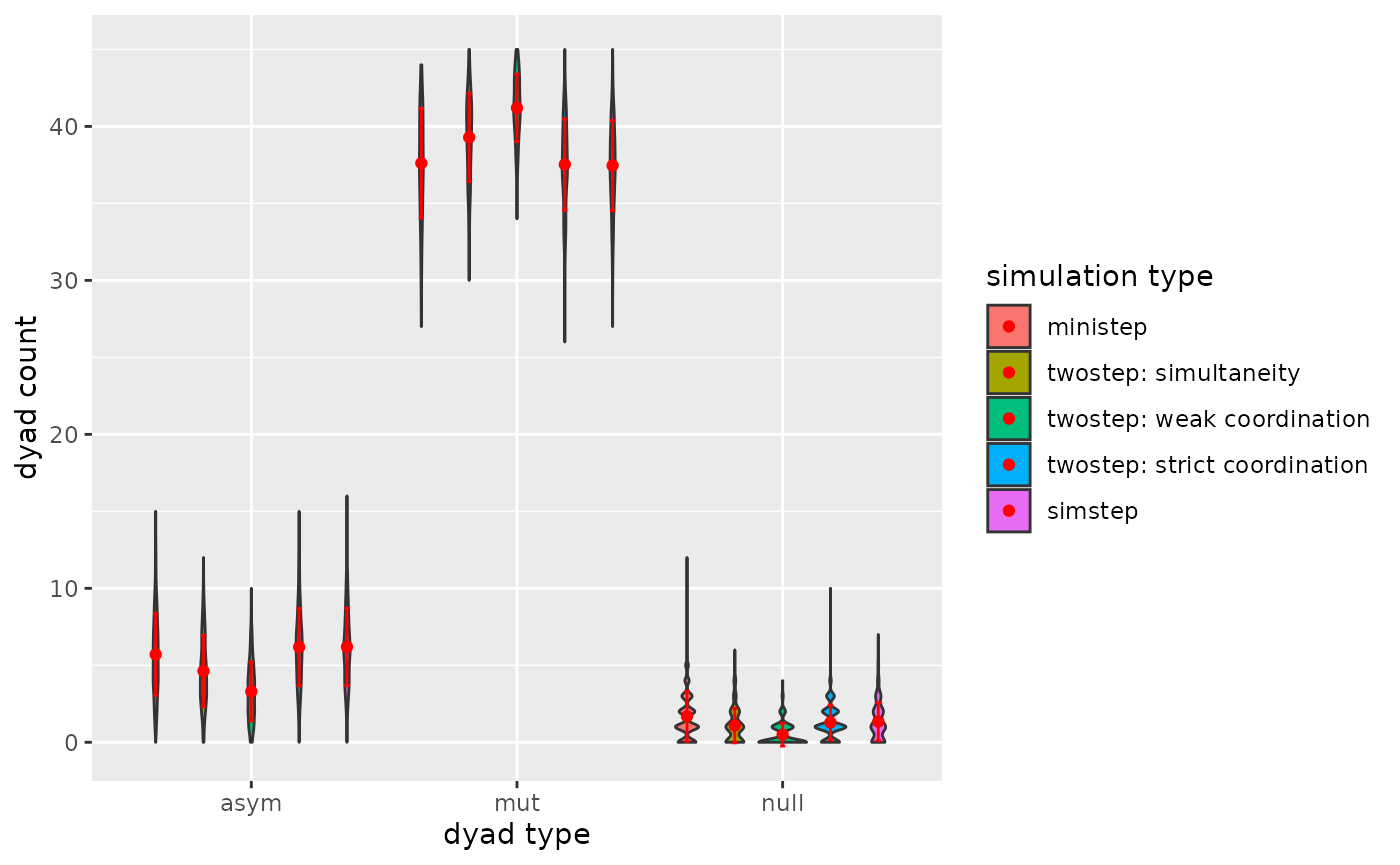
Figure 3.3.3. Dyadcensus Model 3
3.3.4. Counting triads
df1 <- ts_triads(sims=sims1, simtype="ministep")
df2 <- ts_triads(sims=sims2, simtype="twostep: simultaneity")
df3 <- ts_triads(sims=sims3, simtype="twostep: weak coordination")
df4 <- ts_triads(sims=sims4, simtype="twostep: strict coordination")
df5 <- ts_triads(sims=sims5, simtype="simstep")
df <- rbind(df1, df2, df3, df4, df5)3.3.5. Plot results of the triadcensus
dftest <- df[df$x=="030T" | df$x=="120U", ]
p1 <- ggplot(dftest, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep"))))+
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "triad type", y = "triad count", fill="simulation type")
p1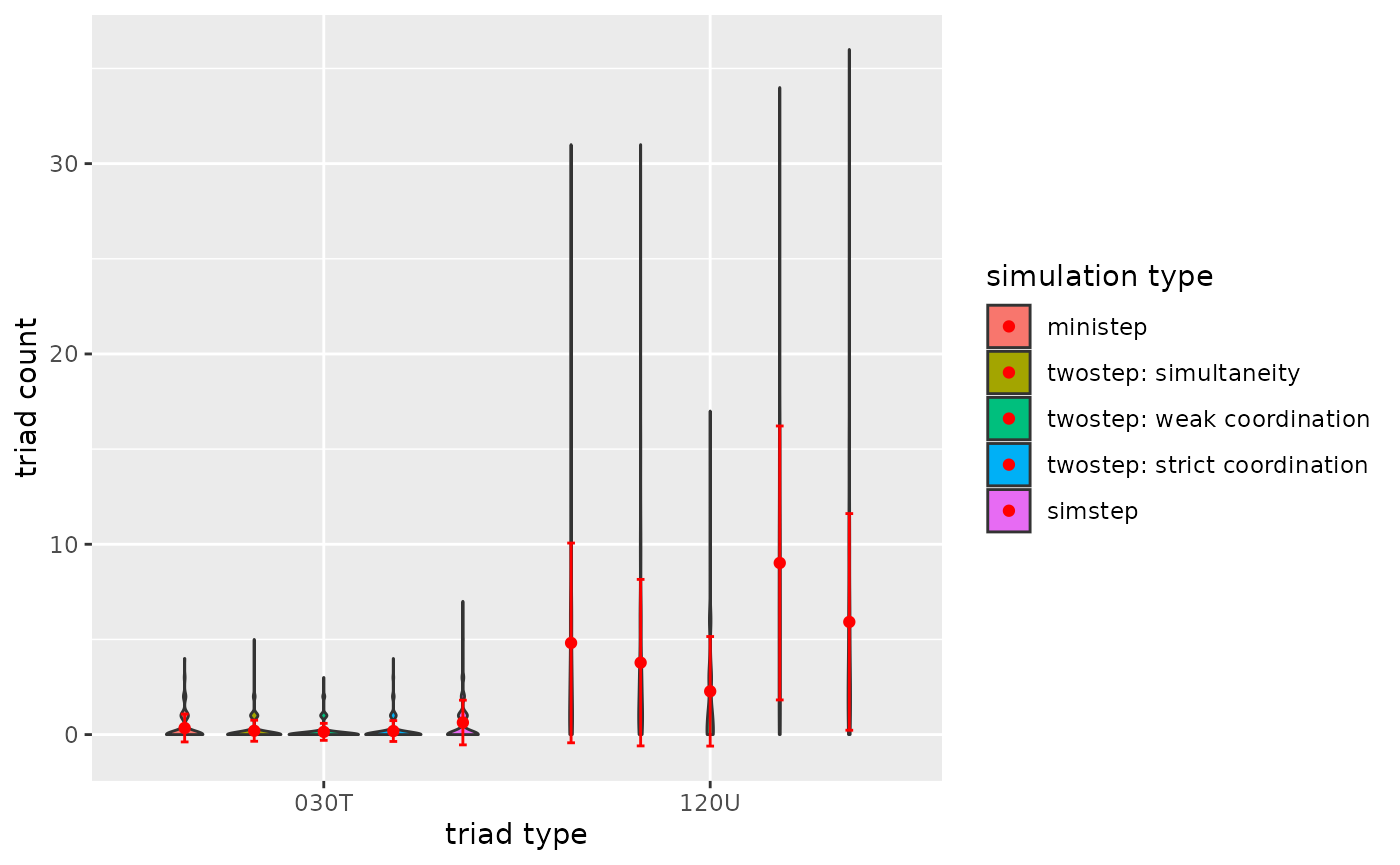
Figure 3.3.5a. Triadcensus Model 3 (Triads 030T & 120U)
dftest <- df[df$x=="120D" | df$x=="120C" , ]
p2 <- ggplot(dftest, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep")))) +
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "triad type", y = "triad count", fill="simulation type")
p2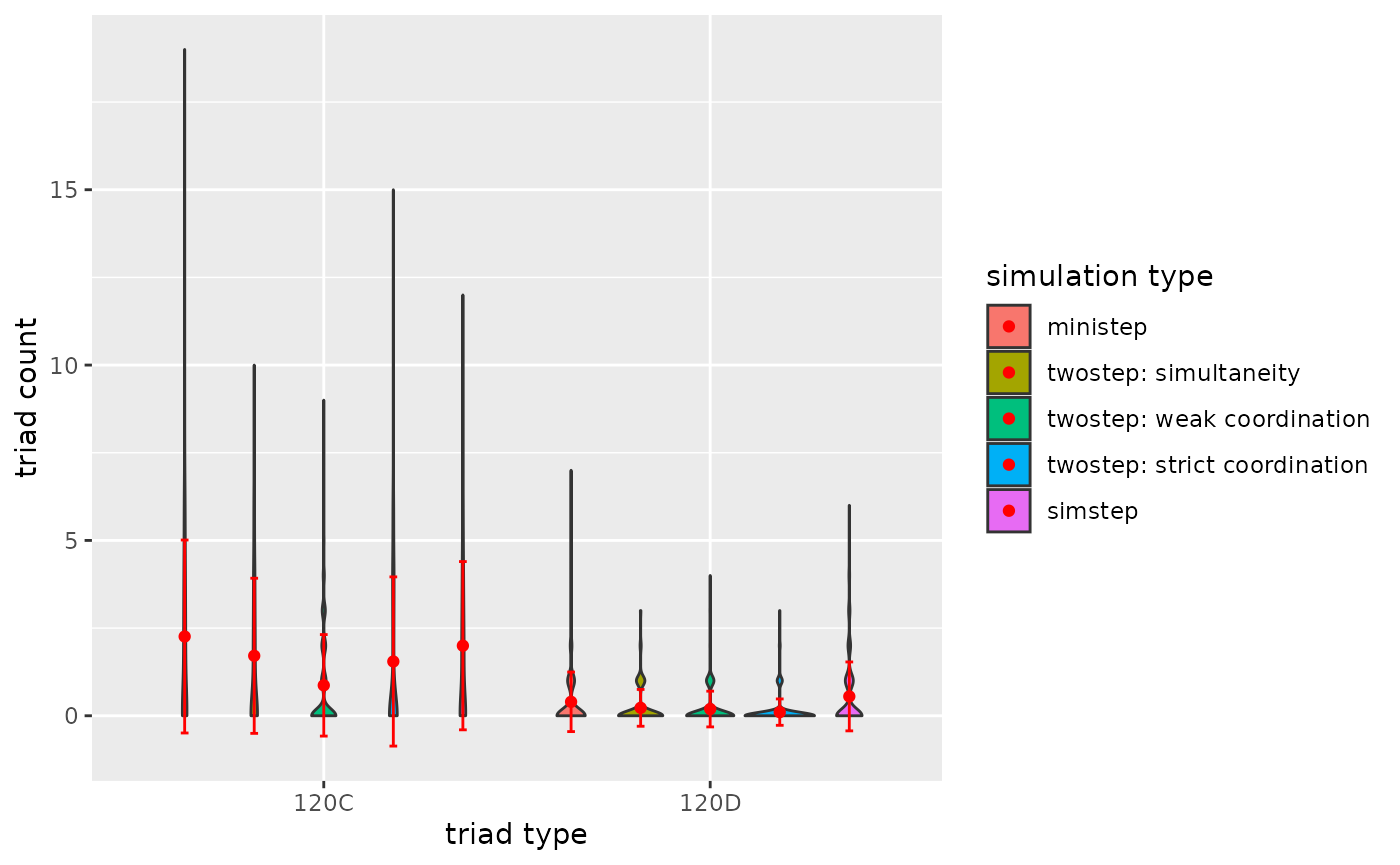
Figure 3.2.5b. Triadcensus Model 3 (Triads 120D & 120C)
dftest <- df[df$x=="210" | df$x=="300" , ]
p3 <- ggplot(dftest, aes(x=x, y=y, fill=factor(type, levels=c("ministep", "twostep: simultaneity", "twostep: weak coordination", "twostep: strict coordination", "simstep")))) +
geom_violin(position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "errorbar",
fun.max = function(x) mean(x) + sd(x),
fun.min = function(x) mean(x) - sd(x),
width=.1,
color="red", position=position_dodge(.9)) +
stat_summary(fun = mean,
geom = "point",
color="red", position=position_dodge(.9)) +
labs(x = "triad type", y = "triad count", fill="simulation type")
p3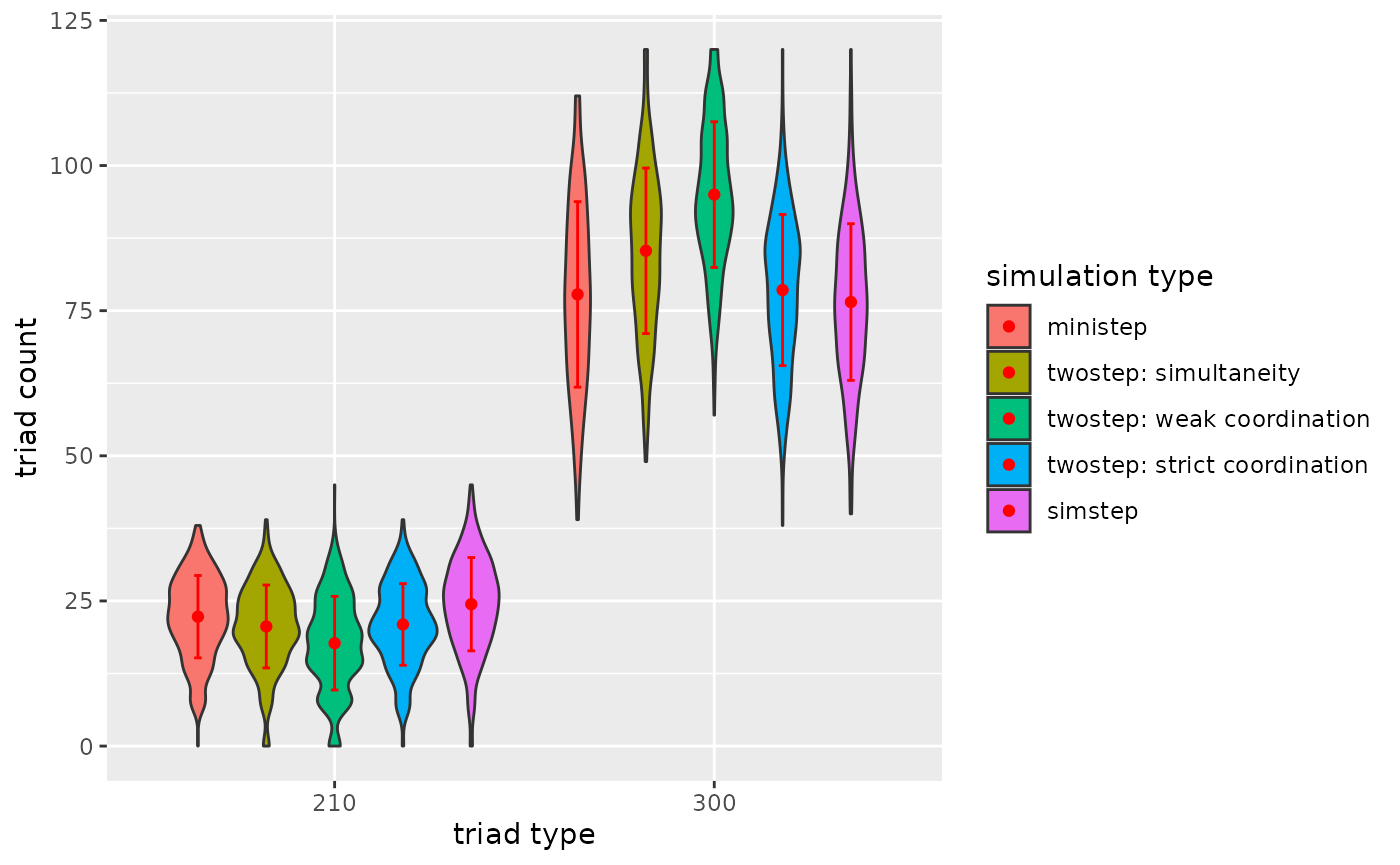
Figure 3.2.5c. Triadcensus Model 3 (Triads 210 & 300)
3.3.6. Conclusion
For this network (ts_net1) and given the network
statistics degree, reciprocity, transitivity and mediated
transitivity…
- It is clearly strict coordination that leads to diverging dyad and
triad counts.
- I tentatively conclude that, as expected, transitive triads are more
common under coordination.
- I tentatively conclude that especially strict coordination leads to transitive triads (except triads 210 and 300).
4. Conclusion
- The different ABMs may give a different dyad and triad count (given
the same start network and network statistics).
- Which ABM leads to the most diverging dyad and triad count depends
on the included network statistics.
- It is difficult to predict (complexity!) how the results of the ABMs will diverge a priori.
